A reference guide for exploratory data analysis (EDA) in Python and R. Genetic data from The Cancer Genome Atlas (TCGA) is used to walk through some routine tasks when you first get your hands on a new (semi-clean) dataset.
Table of Contents
Intro
Motivation
R and Python both have their pros and cons in the data science world. Sometimes I find myself using one language exclusively for an extended period of time, and then switching back to the other often entails some re-learning of basic usage. This tutorial gives side-by-side comparisons of R and Python code to jump-start the relearning process or act as a quick reference guide for those unfamiliar with one language.
For comparisons, code snippets below are posted side-by-side with Python on the left and R on the right. Since the output tables and images are essentially identical by design, only Python output is displayed, but full R and Python code/output can be viewed in the Rmarkdown and Jupyter notebook, respectively, which are available on the GitHub repo.
The Dataset: TCGA
TCGA was a monumental research endeavor to characterize the genetic landscapes of a variety of cancers. Essentially, hundreds of patients had tumor samples profiled for genetic mutations. Most of the data is published online, and we’ll be using some publicly available data for this guide. You don’t have to be too familiar in genetics to follow along with this analysis. We’ll just explore some basic details such as which genes had the most mutations and how many mutations existed per tumor. Simple explanations are given for some genetic terminology.
The data comes in the form of multiple verbose .maf files (detailed
here and
here),
but after some processing we’ll simplify it to a single table looking something like
| GENE | SAMPLE | MUTATION |
|---|---|---|
| A | 1 | \(\alpha\) |
| A | 2 | \(\alpha\) |
| A | 3 | \(\alpha\) |
| B | 1 | \(\beta\) |
| B | 1 | \(\gamma\) |
| B | 2 | \(\beta\) |
| B | 2 | \(\gamma\) |
| C | 1 | \(\delta\) |
| C | 1 | \(\epsilon\) |
| C | 1 | \(\zeta\) |
The elements are just dummy fillers and there will be some other columns, but essentially we are dealing with long-format data and primarily interested in which mutations occurred in which genes and in which samples.
To download the data, first download the GDC Data Transfer Tool and a manifest file, then simply run
./gdc-client download -m MANIFEST.txt
from a terminal. The manifest file used for this analysis contains all
the publicly available data from the breast cancer (BRCA) dataset. However,
the analysis is quite general so you could use any dataset from TCGA that is in
the .maf format.
Notice that the manifest contains multiple .maf files. Essentially, each
refers to a different algorithm used to determine if a mutation exists or
not given genetic sequencing data. The different algorithms and the software
that implements them are typically referred to as callers.
Dependencies
First, let’s load the dependencies.
For Python we make heavy use of the pandas library for data wrangling
plus matplotlib and seaborn (built on top of matplotlib) for visualizations.
The seaborn library changes the default plot styling, and we further
tweak it with some preset themes.
Most of the R packages needed are
contained in the tidyverse mega-package, but cowplot is also useful for
creating a single figure with multiple plots. Further, like seaborn, cowplot
changes the default ggplot2 (part of tidyverse) plot styling.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from collections import OrderedDict
%matplotlib inline
## Plot styling
plt.style.use('seaborn-poster') # Better sizing
plt.style.use('seaborn-white') # White backgroundlibrary(tidyverse)
library(cowplot)Read and clean
Read single file
First, define where the data lives. This is identical in each language (although
you’ll often see people use <- for assignment in R instead of =).
in_path_varscan = 'data/maf/6c93f518-1956-4435-9806-37185266d248/TCGA.BRCA.varscan.6c93f518-1956-4435-9806-37185266d248.DR-10.0.somatic.maf.gz'
in_path_muse = 'data/maf/b8ca5856-9819-459c-87c5-94e91aca4032/TCGA.BRCA.muse.b8ca5856-9819-459c-87c5-94e91aca4032.DR-10.0.somatic.maf.gz'
in_path_ss = 'data/maf/7dd592e3-5950-4438-96d5-3c718aca3f13/TCGA.BRCA.somaticsniper.7dd592e3-5950-4438-96d5-3c718aca3f13.DR-10.0.somatic.maf.gz'
in_path_mutect = 'data/maf/995c0111-d90b-4140-bee7-3845436c3b42/TCGA.BRCA.mutect.995c0111-d90b-4140-bee7-3845436c3b42.DR-10.0.somatic.maf.gz'If we were only interested in a single .maf, reading data would be quite easy.
Just tell the readers where the file is and in this case that the comment character
in .maf is a number symbol #. It is also worth noting that these files are
tab-separated, so we may use the readers for .tsv files.
df = pd.read_table(
in_path_mutect,
comment = '#'
)df = read_tsv(
in_path_mutect,
comment = '#'
)If you run this, you’ll actually get warnings in both languages that some columns have mixed types (e.g. strings and integers). We’ll handle that soon.
Preview
print(df.shape)
df.head(10)print(dim(df))
head(df, 10)Both of the above output dimensions of 120988 x 120 and a short 10-row preview
of the columns (omit the 10 argument to use default values). Only a subset of
the 120 columns are printed here.
| Hugo_Symbol | Entrez_Gene_Id | Center | NCBI_Build | Chromosome | Start_Position | End_Position | Strand | Variant_Classification | Variant_Type | ... | FILTER | CONTEXT | src_vcf_id | tumor_bam_uuid | normal_bam_uuid | case_id | GDC_FILTER | COSMIC | MC3_Overlap | GDC_Validation_Status | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | USP24 | 23358 | WUGSC | GRCh38 | chr1 | 55159655 | 55159655 | + | Missense_Mutation | SNP | ... | panel_of_normals | CTGGATTGTAG | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | common_in_exac;gdc_pon | NaN | True | Unknown |
| 1 | ERICH3 | 127254 | WUGSC | GRCh38 | chr1 | 74571494 | 74571494 | + | Missense_Mutation | SNP | ... | PASS | TTCCTCTACCA | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1474194 | True | Unknown |
| 2 | KIF26B | 55083 | WUGSC | GRCh38 | chr1 | 245419680 | 245419680 | + | Silent | SNP | ... | PASS | GCCTCGCAGGG | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1473725;COSM1473726 | True | Unknown |
| 3 | USP34 | 9736 | WUGSC | GRCh38 | chr2 | 61189055 | 61189055 | + | Silent | SNP | ... | PASS | AAAGCGAGTGC | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1483177 | True | Unknown |
| 4 | ANTXR1 | 84168 | WUGSC | GRCh38 | chr2 | 69245305 | 69245305 | + | Silent | SNP | ... | PASS | TCCTCGCCGCC | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1409122 | True | Unknown |
| 5 | SCN9A | 6335 | WUGSC | GRCh38 | chr2 | 166199365 | 166199365 | + | Silent | SNP | ... | PASS | AGTATGACTGC | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1482144;COSM4814664 | True | Unknown |
| 6 | FN1 | 2335 | WUGSC | GRCh38 | chr2 | 215397809 | 215397809 | + | Nonsense_Mutation | SNP | ... | PASS | CACTTCTCGTG | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1482746;COSM1482747 | True | Unknown |
| 7 | SPHKAP | 80309 | WUGSC | GRCh38 | chr2 | 228016738 | 228016738 | + | Missense_Mutation | SNP | ... | PASS | TCTTTCCTCGG | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1482832;COSM1482833 | True | Unknown |
| 8 | HRH1 | 3269 | WUGSC | GRCh38 | chr3 | 11259653 | 11259653 | + | Missense_Mutation | SNP | ... | PASS | TGCTCATGCTC | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | NaN | COSM1484451 | True | Unknown |
| 9 | LRRC2 | 79442 | WUGSC | GRCh38 | chr3 | 46519054 | 46519054 | + | Missense_Mutation | SNP | ... | panel_of_normals | AGCTGGGAACA | d083d669-6646-463b-853e-c58da8d06439 | 4374e19d-c5e7-49cf-8707-05ae5aeb7369 | aadee87c-6a68-4580-bd10-64ac273b1e3d | 0130d616-885e-4a6c-9d03-2f17dd692a05 | gdc_pon | COSM1485224 | True | Unknown |
Combine multiple files
Now let’s take the .maf from each caller and combine them into a single data
frame. For both languages, the strategy is to create a list of the data frames for each caller
and then concatenate them vertically (i.e. by stacking all the rows on top of each other).
This creates duplicates if a mutation in one sample was
found by multiple callers, so we de-duplicate later.
In most cases, one can probably get away with letting the input readers infer the data type of each column as they are good at guessing, but recall we actually had some warning messages about mixed type columns before. Plus, there was way more data than we need for this exercise. So, below we explicitly define both the columns to read in and their data types.
Note that in R it is often desirable to convert categorical variables/columns from character (string) type to the factor type. Here, the conversion is applied to every character column.
## Define column types
col_type_map = {
'Chromosome': np.unicode,
'Start_Position': np.int64,
'End_Position': np.int64,
'SYMBOL': np.unicode,
'Reference_Allele': np.unicode,
'Allele': np.unicode,
'Variant_Classification': np.unicode,
'IMPACT': np.unicode,
'Variant_Type': np.unicode,
'Tumor_Sample_Barcode': np.unicode,
}
keep_cols = col_type_map.keys()
## Concatenate vertically
in_paths = [
in_path_varscan,
in_path_muse,
in_path_ss,
in_path_mutect
]
df = pd.concat([
pd.read_table(
path,
dtype = col_type_map,
usecols = keep_cols,
comment = '#'
)
for path in in_paths],
ignore_index = True
)## Define column types
col_types = cols_only(
Chromosome = 'c',
Start_Position = 'i',
End_Position = 'i',
SYMBOL = 'c',
Reference_Allele = 'c',
Allele = 'c',
Variant_Classification = 'c',
IMPACT = 'c',
Variant_Type = 'c',
Tumor_Sample_Barcode = 'c'
)
## Concatenate vertically
in_paths = c(
in_path_varscan,
in_path_muse,
in_path_ss,
in_path_mutect
)
df = bind_rows(lapply(
in_paths,
read_tsv,
col_types = col_types,
comment = '#'
))
## Convert characters to factors
df = mutate_if(
df,
is.character,
as.factor
)| Chromosome | Start_Position | End_Position | Variant_Classification | Variant_Type | Reference_Allele | Tumor_Sample_Barcode | Allele | SYMBOL | IMPACT | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1916819 | 1916819 | Missense_Mutation | SNP | C | TCGA-A2-A3Y0-01A-11D-A23C-09 | G | CALML6 | MODERATE |
| 1 | chr1 | 2172304 | 2172304 | Missense_Mutation | SNP | G | TCGA-A2-A3Y0-01A-11D-A23C-09 | C | PRKCZ | MODERATE |
| 2 | chr1 | 3766586 | 3766586 | Missense_Mutation | SNP | G | TCGA-A2-A3Y0-01A-11D-A23C-09 | A | CCDC27 | MODERATE |
| 3 | chr1 | 6040634 | 6040634 | Silent | SNP | G | TCGA-A2-A3Y0-01A-11D-A23C-09 | C | KCNAB2 | LOW |
| 4 | chr1 | 23961791 | 23961791 | Missense_Mutation | SNP | A | TCGA-A2-A3Y0-01A-11D-A23C-09 | G | PNRC2 | MODERATE |
Rename and reorder columns
We essentially create a map between the old and new column names and apply that
to the data frames in both languages. In Python, notice that we’re using an
OrderedDict collection to preserve the order of the columns as we want them
to appear.
## Rename
col_name_map = OrderedDict([
('Chromosome', 'CHR'),
('Start_Position', 'START'),
('End_Position', 'END'),
('SYMBOL', 'GENE'),
('Reference_Allele', 'REF'),
('Allele', 'ALT'),
('Variant_Classification', 'CLASS'),
('IMPACT', 'IMPACT'),
('Variant_Type', 'TYPE'),
('Tumor_Sample_Barcode', 'BARCODE'),
])
df.rename(columns=col_name_map, inplace=True)
## Reorder
keep_cols = list(col_name_map.values()) # Need list() because its odict
df = df[keep_cols]## Rename
col_name_map = list(
CHR = 'Chromosome',
START = 'Start_Position',
END = 'End_Position',
GENE = 'SYMBOL',
REF = 'Reference_Allele',
ALT = 'Allele',
CLASS = 'Variant_Classification',
IMPACT = 'IMPACT',
TYPE = 'Variant_Type',
BARCODE = 'Tumor_Sample_Barcode'
)
df = rename(df, !!! col_name_map)
## Reorder
keep_cols = names(col_name_map)
df = select(df, keep_cols)| CHR | START | END | GENE | REF | ALT | CLASS | IMPACT | TYPE | BARCODE | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1916819 | 1916819 | CALML6 | C | G | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 |
| 1 | chr1 | 2172304 | 2172304 | PRKCZ | G | C | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 |
| 2 | chr1 | 3766586 | 3766586 | CCDC27 | G | A | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 |
| 3 | chr1 | 6040634 | 6040634 | KCNAB2 | G | C | Silent | LOW | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 |
| 4 | chr1 | 23961791 | 23961791 | PNRC2 | A | G | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 |
Fill Missing Data
Some of the GENE column values are set to the none/NA/missing value in each
language. This is reasonably accurate as the mutations occur between genes, but
there is a more descriptive technical term: “intergenic” mutations. Here, we
reassign all the missing GENE values to “INTERGENIC”.
Since GENE is a factor in the R dataframe, we have to add the new value to the
existing levels before reassignment.
df.fillna(
{'GENE': 'INTERGENIC'},
inplace = True
)levels(df$GENE) = c(levels(df$GENE), 'INTERGENIC')
df$GENE[is.na(df$GENE)] = 'INTERGENIC'New columns from string operations
We create a new SAMPLE column by selecting the first 12 characters from the
BARCODE column e.g. barcode TCGA-A2-A3Y0-01A-11D-A23C-09 refers to sample
TCGA-A2-A3Y0. The rest of the barcode references other properties we are
not concerned with here (such as if the sample is a metastatic or primary tumor).
Further, we create a unique identifier for each mutation by joining the CHR,
GENE, START, END, REF, and ALT columns together with a colon delimiter.
That is, if a mutation changes the C nucleotide at position 1916819 on chromosome 1
to a G, then the mutation is in the CALML6 gene, and we refer to it as
chr1:CALML6:1916819:1916819:C:G. Note that the start and end positions will be
different if we are dealing with an insertion or deletion of nucleotides rather
than a single-nucleotide polymorphism (SNP).
Finally, a TYPE2 column is created which condenses the information from the
TYPE column. TYPE tells if the mutation is a “SNP” (single nucleotide polymorphism
e.g. one nucleotide changes to another), “INS” (insertion), or “DEL” (deletion).
Insertions and deletions insert or delete one or more nucleotides, and they are
often collectively referred to as “indels”. The TYPE2 column will contain
only “SNP” and “INDEL” values.
Similar to when we read the data, if necessary we should convert character variable to factor variables in R.
Also in R, there are 2 common methods to accomplish
the above tasks, both worth noting. One is to create the columns directly by assignment
as done in Python, and the other is to use the mutate() function from the dplyr
package. Both are basically the same, but mutate() has the advantage of not
having to constantly reference columns by the $ sign.
df['SAMPLE'] = df['BARCODE'].str[0:12]
df['MUTATION'] = df['CHR'].str.cat([
df['GENE'],
df['START'].map(str),
df['END'].map(str),
df['REF'],
df['ALT']],
sep = ':'
)
df['TYPE2'] = ['SNP' if TYPE == 'SNP' else 'INDEL' for TYPE in df['TYPE']]## New columns by assignment
df$SAMPLE = str_sub(df$BARCODE, 1, 12)
df$MUTATION = paste(
df$CHR,
df$GENE,
df$START,
df$END,
df$REF,
df$ALT,
sep = ':'
)
df$TYPE2 = ifelse(df$TYPE == 'SNP', 'SNP', 'INDEL')
## New columns with mutate()
# Equivalent to above (and thus redundant)
df = mutate(
df,
SAMPLE = str_sub(BARCODE, 1, 12),
MUTATION = paste(
CHR,
GENE,
START,
END,
REF,
ALT,
sep = ':'
),
TYPE2 = ifelse(TYPE == 'SNP', 'SNP', 'INDEL')
)
## Convert new columns to factors if applicable,
## possibly with manually-set levels
df$SAMPLE = as.factor(df$SAMPLE)
df$MUTATION = as.factor(df$MUTATION)
df$TYPE2 = factor(df$TYPE2, levels = c('SNP', 'INDEL'))| CHR | START | END | GENE | REF | ALT | CLASS | IMPACT | TYPE | BARCODE | SAMPLE | MUTATION | TYPE2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1916819 | 1916819 | CALML6 | C | G | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:CALML6:1916819:1916819:C:G | SNP |
| 1 | chr1 | 2172304 | 2172304 | PRKCZ | G | C | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:PRKCZ:2172304:2172304:G:C | SNP |
| 2 | chr1 | 3766586 | 3766586 | CCDC27 | G | A | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:CCDC27:3766586:3766586:G:A | SNP |
| 3 | chr1 | 6040634 | 6040634 | KCNAB2 | G | C | Silent | LOW | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:KCNAB2:6040634:6040634:G:C | SNP |
| 4 | chr1 | 23961791 | 23961791 | PNRC2 | A | G | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:PNRC2:23961791:23961791:A:G | SNP |
Remove duplicates
Since we generated duplicate rows when combining data frames for each caller, we remove them so they don’t interfere with counting and statistics. Since the rows would be exactly identical for each caller, we can just look for where entire rows are duplicated.
Further, it might be useful to have a dataframe with only mutation information and
without reference to which samples were found. That is, one row per mutation instead
of one row per-mutation per-sample. For this, the deduplication process should
consider duplicate values in the MUTATION column only.
## Remove duplicate rows
df.drop_duplicates(inplace = True)
## Remove duplicate based on column(s)
dupe_cols = ['MUTATION']
df_mut = df.drop_duplicates(dupe_cols)## Remove duplicate rows
df = unique(df)
## Remove duplicate based on column
df_mut = df[!duplicated(df$MUTATION),]By checking the shape of each frame, we see that df has shape 132916 x 13 and df_mut is 130625 x 13.
Remove columns
The df_mut dataframe above still contains sample information, but it should only
have mutation information. Thus, the SAMPLE and BARCODE columns should be dropped.
Note that with pandas in Python you might encounter a warning about chaining
operations (e.g. reading -> renaming -> dedupe -> drop columns) resulting in
unintended effects. You can
read about why the issue arises
and suppress the warning if you feel comfortable
about chaining operations.
drop_cols = ['BARCODE', 'SAMPLE']
df_mut.drop(
columns = drop_cols,
inplace = True
)drop_cols = c('BARCODE', 'SAMPLE')
df_mut = select(df_mut, -one_of(drop_cols))| CHR | START | END | GENE | REF | ALT | CLASS | IMPACT | TYPE | MUTATION | TYPE2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1916819 | 1916819 | CALML6 | C | G | Missense_Mutation | MODERATE | SNP | chr1:CALML6:1916819:1916819:C:G | SNP |
| 1 | chr1 | 2172304 | 2172304 | PRKCZ | G | C | Missense_Mutation | MODERATE | SNP | chr1:PRKCZ:2172304:2172304:G:C | SNP |
| 2 | chr1 | 3766586 | 3766586 | CCDC27 | G | A | Missense_Mutation | MODERATE | SNP | chr1:CCDC27:3766586:3766586:G:A | SNP |
| 3 | chr1 | 6040634 | 6040634 | KCNAB2 | G | C | Silent | LOW | SNP | chr1:KCNAB2:6040634:6040634:G:C | SNP |
| 4 | chr1 | 23961791 | 23961791 | PNRC2 | A | G | Missense_Mutation | MODERATE | SNP | chr1:PNRC2:23961791:23961791:A:G | SNP |
New columns from delimited column
Here we essentially reverse the process of making the mutation column. In other words, assume
we start with a MUTATION column that has values like chr1:CALML6:1916819:1916819:C:G.
The goal is to create six new columns with values like chr1, CALM6, etc.
Since we already have those columns in the existing df, first we make a dummy
frame without them so we can verify that the procedure works.
## Create dummy frame
df_tmp = df.copy()
drop_cols = [
'CHR',
'GENE',
'START',
'END',
'REF',
'ALT'
]
df_tmp.drop(columns = drop_cols, inplace = True)
## Create new columns
(df_tmp['CHR'], df_tmp['GENE'], df_tmp['START'],
df_tmp['END'], df_tmp['REF'], df_tmp['ALT']) = (
df_tmp['MUTATION'].str.split(':').str )## Create dummy frame
df_tmp = df
drop_cols = c(
'CHR',
'GENE',
'START',
'END',
'REF',
'ALT'
)
df_tmp = select(df_tmp, -one_of(drop_cols))
## Create new columns
df_tmp = separate(
df_tmp,
MUTATION,
drop_cols,
':'
)| CLASS | IMPACT | TYPE | BARCODE | SAMPLE | MUTATION | TYPE2 | CHR | GENE | START | END | REF | ALT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:CALML6:1916819:1916819:C:G | SNP | chr1 | CALML6 | 1916819 | 1916819 | C | G |
| 1 | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:PRKCZ:2172304:2172304:G:C | SNP | chr1 | PRKCZ | 2172304 | 2172304 | G | C |
| 2 | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:CCDC27:3766586:3766586:G:A | SNP | chr1 | CCDC27 | 3766586 | 3766586 | G | A |
| 3 | Silent | LOW | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:KCNAB2:6040634:6040634:G:C | SNP | chr1 | KCNAB2 | 6040634 | 6040634 | G | C |
| 4 | Missense_Mutation | MODERATE | SNP | TCGA-A2-A3Y0-01A-11D-A23C-09 | TCGA-A2-A3Y0 | chr1:PNRC2:23961791:23961791:A:G | SNP | chr1 | PNRC2 | 23961791 | 23961791 | A | G |
Write files
df.to_csv(
'data/out/df.tsv',
sep = '\t'
)write_delim(
df,
'data/out/df_r.tsv',
delim = '\t'
)Read and write binary files
Both R and Python have methods to read/write binary data for efficiency.
pickle_path = 'data/out/df.pkl'
# Write
df.to_pickle(pickle_path)
# Read
df = pd.read_pickle(pickle_path)rds_path = 'data/out/df.rds'
## Write
saveRDS(df, rds_path)
## Read
df = readRDS(rds_path)Long to wide
The goal here is to go from
| GENE | SAMPLE | MUTATION |
|---|---|---|
| A | 1 | \(\alpha\) |
| A | 2 | \(\alpha\) |
| A | 3 | \(\alpha\) |
| B | 1 | \(\beta\) |
| B | 1 | \(\gamma\) |
| B | 2 | \(\beta\) |
| B | 2 | \(\gamma\) |
| C | 1 | \(\delta\) |
| C | 1 | \(\epsilon\) |
| C | 1 | \(\zeta\) |
to
| GENE | MUTATION | 1 | 2 | 3 |
|---|---|---|---|---|
| A | \(\alpha\) | 1 | 1 | 1 |
| B | \(\beta\) | 1 | 1 | 0 |
| B | \(\gamma\) | 1 | 1 | 0 |
| C | \(\delta\) | 0 | 0 | 1 |
| C | \(\epsilon\) | 0 | 0 | 1 |
| C | \(\zeta\) | 0 | 0 | 1 |
That is, make each row correspond to a mutation and create columns for each sample. The sample columns are filled with 1 if the mutation exists and 0 otherwise. This transformation is also knowing as casting.
If you’re familiar with genetics / bioinformatics, the wide format is similar to how a VCF file is structured while MAF structure is long.
# Create a dummy column w/ fill value
df_tmp = df.copy()
df_tmp['EXISTS'] = 1
# Long-to-wide
df_wide = df_tmp.pivot(
index = 'MUTATION',
columns = 'SAMPLE',
values = 'EXISTS'
)
del df_tmp
## Clean-up column names
df_wide.columns = df_wide.columns.values
df_wide.reset_index(inplace = True)
## Fill empty values
df_wide.fillna(0, inplace = True)## Create a dummy column w/ fill value
df_tmp = select(df, c('SAMPLE', 'MUTATION'))
df_tmp$EXISTS = 1
## Long-to-wide
df_wide = spread(
df_tmp,
key = 'SAMPLE',
value = 'EXISTS',
fill = 0
)
rm(df_tmp)Note that df_wide in both cases does not contain columns like GENE.
Additional columns are added below. Also, only a subset of the sample columns
are shown below (and likely they are all filled with 0).
| MUTATION | TCGA-3C-AAAU | TCGA-3C-AALI | TCGA-3C-AALJ | TCGA-3C-AALK | TCGA-4H-AAAK | TCGA-5L-AAT0 | TCGA-5L-AAT1 | TCGA-5T-A9QA | TCGA-A1-A0SD | ... | TCGA-UL-AAZ6 | TCGA-UU-A93S | TCGA-V7-A7HQ | TCGA-W8-A86G | TCGA-WT-AB41 | TCGA-WT-AB44 | TCGA-XX-A899 | TCGA-XX-A89A | TCGA-Z7-A8R5 | TCGA-Z7-A8R6 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr10:A1CF:50813906:50813906:G:- | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | chr10:A1CF:50813932:50813932:G:T | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | chr10:A1CF:50828193:50828193:C:A | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | chr10:A1CF:50836094:50836094:G:A | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | chr10:A1CF:50836177:50836177:G:A | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Merge
Add the GENE and IMPACT columns that exist in one dataframe (df_mut) to the
newly created df_wide which only has MUTATION and sample columns.
## Select columns to add on and col to join on
cols = ['MUTATION', 'GENE', 'IMPACT']
## Merge
df_wide = pd.merge(
df_wide, df_mut[cols],
on = 'MUTATION',
how = 'left',
)
## Re-order columns
# Put new columns first
cols = df_wide.columns.tolist()
cols = cols[-2:] + cols[:-2]
df_wide = df_wide[cols]## Select columns to add on and col to join on
cols = c('MUTATION', 'GENE', 'IMPACT')
## Merge
df_wide = merge(
df_wide, select(df_mut, cols),
by = 'MUTATION',
all.x = TRUE
)
## Re-order columns
# Put new columns first
df_wide = select(df_wide, cols, everything())Again, only a subset of the sample columns are shown (probably all filled with 0).
| GENE | IMPACT | MUTATION | TCGA-3C-AAAU | TCGA-3C-AALI | TCGA-3C-AALJ | TCGA-3C-AALK | TCGA-4H-AAAK | TCGA-5L-AAT0 | TCGA-5L-AAT1 | ... | TCGA-UL-AAZ6 | TCGA-UU-A93S | TCGA-V7-A7HQ | TCGA-W8-A86G | TCGA-WT-AB41 | TCGA-WT-AB44 | TCGA-XX-A899 | TCGA-XX-A89A | TCGA-Z7-A8R5 | TCGA-Z7-A8R6 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A1CF | HIGH | chr10:A1CF:50813906:50813906:G:- | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | A1CF | MODERATE | chr10:A1CF:50813932:50813932:G:T | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | A1CF | MODERATE | chr10:A1CF:50828193:50828193:C:A | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | A1CF | MODERATE | chr10:A1CF:50836094:50836094:G:A | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | A1CF | LOW | chr10:A1CF:50836177:50836177:G:A | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Wide to long
We invert the procedure from long to wide (this is also known as melting). Note that for this particular dataset, the operation is quite memory intensive.
# Set value and id columns
value_mask = df_wide.columns.str.startswith('TCGA-')
value_cols = df_wide.columns[value_mask]
id_cols = ['GENE', 'IMPACT', 'MUTATION']
# Melt
df_long = pd.melt(
df_wide,
id_cols,
value_cols,
var_name = 'SAMPLE',
value_name = 'EXISTS'
)
# Clean up
df_long = df_long.query('EXISTS != 0')
df_long.drop(columns = 'EXISTS', inplace = True)# Set value and id columns
value_cols = grep('TCGA-', names(df_wide), value = TRUE)
# Melt
df_long = gather(
df_wide,
key = 'SAMPLE',
value = 'EXISTS',
value_cols
)
# Clean up
df_long = filter(df_long, EXISTS != 0)
df_long = select(df_long, -EXISTS)| GENE | IMPACT | MUTATION | SAMPLE | |
|---|---|---|---|---|
| 967 | CELF2 | MODIFIER | chr10:CELF2:11333805:11333805:A:- | TCGA-3C-AAAU |
| 1972 | GATA3 | HIGH | chr10:GATA3:8073911:8073912:-:A | TCGA-3C-AAAU |
| 4782 | WDR11 | MODIFIER | chr10:WDR11:120909443:120909443:G:A | TCGA-3C-AAAU |
| 6124 | CD248 | MODERATE | chr11:CD248:66314996:66314996:C:T | TCGA-3C-AAAU |
| 8170 | MALAT1 | MODIFIER | chr11:MALAT1:65505435:65505437:AAA:- | TCGA-3C-AAAU |
Free memory
del df_wide, df_longrm(df_wide, df_long)Counting and summaries
Various descriptive calculations are done with the data now in the proper format. These calculations are often paired with graphic representations which are covered in the next section.
Count unique elements
n_genes = df['GENE'].nunique()
n_samples = df['BARCODE'].nunique()
n_mutations = df['MUTATION'].nunique()
n_mutation_classes = df['CLASS'].nunique()n_genes = length(unique(df$GENE))
n_samples = length(unique(df$SAMPLE))
n_mutations = length(unique(df$MUTATION))
n_mutation_classes = length(unique(df$CLASS))The values are
Number of unique Samples: 986 Genes: 19168 Mutations: 130625 Mutation Classes: 18
Count by factor levels
From the above, we saw that, for example, there are 18 mutation classes such as
missense mutations, silent mutations, etc. What would probably be more useful is
to know how many mutations are there of each class. Similarly, for the IMPACT
column, it would be useful to know how many HIGH impact, LOW impact, etc. mutations
were found.
class_counts = df['CLASS'].value_counts()
impact_counts = df['IMPACT'].value_counts()
type_counts = df['TYPE'].value_counts()
mut_counts = df['MUTATION'].value_counts()class_counts = sort(summary(df$CLASS), decreasing = TRUE)
impact_counts = sort(summary(df$IMPACT), decreasing = TRUE)
type_counts = sort(summary(df$TYPE), decreasing = TRUE)
# Need to override the maxsum = 50 arg
mut_counts = sort(
summary(df$MUTATION, maxsum = n_mutations),
decreasing = TRUE)A quick preview of the counts are
Counts per CLASS: Missense_Mutation 66371 Silent 23881 3'UTR 11052 Intron 6990 Nonsense_Mutation 6056 Frame_Shift_Del 3661 5'UTR 3392 RNA 2543 Frame_Shift_Ins 1975 Splice_Site 1921 Splice_Region 1537 3'Flank 1132 5'Flank 906 In_Frame_Del 869 In_Frame_Ins 433 Nonstop_Mutation 93 Translation_Start_Site 90 IGR 14 Name: CLASS, dtype: int64 Counts per IMPACT: MODERATE 67648 MODIFIER 26059 LOW 25413 HIGH 13796 Name: IMPACT, dtype: int64 Counts per TYPE: SNP 121319 DEL 7316 INS 4281 Name: TYPE, dtype: int64 Top repeated (>10) mutations: chr3:PIK3CA:179234297:179234297:A:G 121 chr3:PIK3CA:179218303:179218303:G:A 63 chr3:PIK3CA:179218294:179218294:G:A 43 chr1:ST6GALNAC3:76576946:76576947:-:AAAC 33 chr14:AKT1:104780214:104780214:C:T 25 chr3:MUC4:195783009:195783009:C:T 21 chr10:GATA3:8069470:8069471:CA:- 21 chr3:MUC4:195783008:195783008:A:G 20 chr17:TP53:7675088:7675088:C:T 20 chr3:PIK3CA:179203765:179203765:T:A 17 chr15:GOLGA6L6:20535018:20535018:C:T 17 chr6:OPRM1:154107953:154107958:TTTTTA:- 13 chr3:PIK3CA:179234297:179234297:A:T 13 chr17:TP53:7673802:7673802:C:T 12 chr1:NBPF12:146963189:146963189:G:C 11 chr16:PKD1P6:15104542:15104542:T:A 11 Name: MUTATION, dtype: int64 Samples with most mutations: TCGA-AN-A046 7948 TCGA-AC-A23H 6711 TCGA-5L-AAT1 2117 TCGA-BH-A18G 2033 TCGA-AN-A0AK 2029 TCGA-A8-A09Z 1924 TCGA-BH-A0HF 1695 TCGA-AO-A128 1644 TCGA-D8-A1XK 1541 TCGA-BH-A0B6 1419 Name: SAMPLE, dtype: int64 Samples with least mutations: TCGA-A2-A0ES 18 TCGA-AC-A2FB 17 TCGA-AR-A24W 16 TCGA-LL-A440 16 TCGA-AR-A252 16 TCGA-AO-A1KO 15 TCGA-A2-A1G6 12 TCGA-A8-A08C 9 TCGA-A2-A25F 7 TCGA-AC-A2FK 2 Name: SAMPLE, dtype: int64
Summarize by group
Consider the toy example dataframe again:
| GENE | SAMPLE | MUTATION |
|---|---|---|
| A | 1 | \(\alpha\) |
| A | 2 | \(\alpha\) |
| A | 3 | \(\alpha\) |
| B | 1 | \(\beta\) |
| B | 1 | \(\gamma\) |
| B | 2 | \(\beta\) |
| B | 2 | \(\gamma\) |
| C | 1 | \(\delta\) |
| C | 1 | \(\epsilon\) |
| C | 1 | \(\zeta\) |
For quantifying the number of mutations, we might be interested in any of the following three metrics:
- Genes with the most samples mutated
- Genes with the largest number of aggregate mutations
- Genes with the largest number of unique mutations
The counts for the above example are as follows (bolded cells are the highest count for the column):
| GENE | # Samples Mutated | # Aggregate Mutations | # Unique Mutations |
|---|---|---|---|
| A | 3 | 3 | 1 |
| B | 2 | 4 | 2 |
| C | 1 | 3 | 3 |
All three metrics are calculated below and saved into a dataframe (df_summary) like the above
example.
Note that by default in the Python example, the dataframe columns will be a MultiIndex which is somewhat cumbersome for this simple task, so some extra work is done to simplify it to what we see above and get by default in R.
## Summarize by group
df_summary = df.groupby('GENE').agg(OrderedDict([
('MUTATION', ['size', 'nunique']),
('SAMPLE', ['nunique']),
]))
## Rename the MultiIndex cols
# This is why OrderedDict was used w/ agg()
df_summary.columns = [
'N_MUTATIONS',
'N_UNIQUE_MUTATIONS',
'N_SAMPLES'
]
## Make GENE index into a column
df_summary.reset_index(inplace = True)df_summary = df %>%
group_by(GENE) %>%
summarise(
N_MUTATIONS = n(),
N_UNIQUE_MUTATIONS = n_distinct(MUTATION),
N_SAMPLES = n_distinct(BARCODE)
)| GENE | N_MUTATIONS | N_UNIQUE_MUTATIONS | N_SAMPLES | |
|---|---|---|---|---|
| 0 | A1BG | 5 | 5 | 5 |
| 1 | A1CF | 10 | 10 | 10 |
| 2 | A2M | 15 | 15 | 14 |
| 3 | A2ML1 | 15 | 15 | 13 |
| 4 | A4GALT | 2 | 2 | 2 |
| 5 | A4GNT | 3 | 3 | 3 |
| 6 | AAAS | 4 | 4 | 4 |
| 7 | AACS | 7 | 7 | 7 |
| 8 | AACSP1 | 1 | 1 | 1 |
| 9 | AADAC | 6 | 6 | 6 |
Sorting
Now that we have our summary dataframe, often we might be interested in sorting it by one or more columns.
df_summary.sort_values(
['N_SAMPLES', 'N_MUTATIONS'],
ascending = [False, True],
inplace = True
)df_summary = df_summary %>%
arrange(desc(N_SAMPLES), N_MUTATIONS)| GENE | N_MUTATIONS | N_UNIQUE_MUTATIONS | N_SAMPLES | |
|---|---|---|---|---|
| 17100 | TP53 | 377 | 240 | 360 |
| 12150 | PIK3CA | 384 | 82 | 339 |
| 17553 | TTN | 472 | 472 | 243 |
| 10153 | MUC4 | 280 | 141 | 197 |
| 2831 | CDH1 | 154 | 133 | 149 |
Summarize by multiple groups
It is simple to further refine our grouping. Here, rather than just finding
how many mutations are in each gene, we’ll investigate how many HIGH-impact, LOW-impact,
etc. mutations are found in each gene. In other words, we group by both the GENE
and IMPACT columns before counting. Doing so is a straightforward extension of
grouping by a single column.
df_impact = df.groupby(['GENE', 'IMPACT']).agg({
'MUTATION': 'nunique'
})
df_impact.reset_index(inplace = True)df_impact = df %>%
group_by(GENE, IMPACT) %>%
summarise(COUNT = n_distinct(MUTATION))| GENE | IMPACT | MUTATION | |
|---|---|---|---|
| 0 | A1BG | MODERATE | 3 |
| 1 | A1BG | MODIFIER | 2 |
| 2 | A1CF | HIGH | 1 |
| 3 | A1CF | LOW | 1 |
| 4 | A1CF | MODERATE | 6 |
Descriptive statistics
Here classic descriptive stats such as median, upper quartile, minimum, etc. number
of mutations per sample are calculated.
The stats are calculated per TYPE2 per IMPACT. So, before calculating, we
first make a data frame (df_sample_counts) that is appropriately grouped and
counts the number of mutations per grouping. The stats are then compiled in a
second data frame (df_sample_stats).
Note that for visual appeal when creating plots with these numbers later on, we arbitrarily only consider groupings with between 20 and 200 mutations.
## Make counts per group
df_sample_counts = df.groupby(['SAMPLE', 'TYPE2', 'IMPACT']).agg({'MUTATION': 'size'})
df_sample_counts.reset_index(inplace=True)
df_sample_counts.columns = ['SAMPLE', 'TYPE2', 'IMPACT', 'N_MUTATIONS']
df_sample_counts = df_sample_counts.loc[df_sample_counts['N_MUTATIONS'].between(20, 200)]
def quantile(n):
'''
Quantile calculation for use in Pandas .agg()
'''
def quantile_(x):
return pd.Series.quantile(x, n)
quantile_.__name__ = 'percentile_%s' % n
return quantile_
## Calculate stats
df_sample_stats = df_sample_counts.groupby(['TYPE2', 'IMPACT']).agg({
'N_MUTATIONS': [
'median',
quantile(0.75),
quantile(.25),
'mean',
'std',
'min',
'max']
})
## Clean up column names
df_sample_stats.reset_index(inplace = True)
df_sample_stats.columns = [
'TYPE2',
'IMPACT',
'MEDIAN',
'UPPER',
'LOWER',
'MEAN',
'STD',
'MIN',
'MAX']## Make counts per group
df_sample_counts = df %>%
group_by(SAMPLE, TYPE2, IMPACT) %>%
summarize(N_MUTATIONS = n()) %>%
filter(N_MUTATIONS >= 20, N_MUTATIONS <= 200)
## Calculate stats
df_sample_stats = df_sample_counts %>%
group_by(TYPE2, IMPACT) %>%
summarize(
MEDIAN = median(N_MUTATIONS),
UPPER = quantile(N_MUTATIONS, .75),
LOWER = quantile(N_MUTATIONS, .25),
MEAN = mean(N_MUTATIONS),
STD = sd(N_MUTATIONS),
MIN = min(N_MUTATIONS),
MAX = max(N_MUTATIONS)
)df_sample_counts looks like
| SAMPLE | TYPE2 | IMPACT | N_MUTATIONS | |
|---|---|---|---|---|
| 5 | TCGA-3C-AAAU | SNP | MODIFIER | 22 |
| 7 | TCGA-3C-AALI | INDEL | MODIFIER | 21 |
| 8 | TCGA-3C-AALI | SNP | HIGH | 43 |
| 9 | TCGA-3C-AALI | SNP | LOW | 153 |
| 17 | TCGA-3C-AALJ | SNP | MODERATE | 27 |
and df_sample_stats looks like
| TYPE2 | IMPACT | MEDIAN | UPPER | LOWER | MEAN | STD | MIN | MAX | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | INDEL | HIGH | 57.0 | 93.00 | 26 | 69.764706 | 53.634794 | 20 | 196 |
| 1 | INDEL | MODERATE | 49.0 | 68.00 | 36 | 61.800000 | 41.865260 | 25 | 131 |
| 2 | INDEL | MODIFIER | 58.0 | 119.00 | 39 | 78.692308 | 55.741045 | 21 | 173 |
| 3 | SNP | HIGH | 34.5 | 64.75 | 24 | 44.431818 | 27.942131 | 20 | 150 |
| 4 | SNP | LOW | 31.0 | 44.00 | 23 | 39.250000 | 25.638295 | 20 | 157 |
| 5 | SNP | MODERATE | 40.5 | 65.00 | 28 | 52.778364 | 34.457118 | 20 | 190 |
| 6 | SNP | MODIFIER | 30.5 | 45.75 | 24 | 40.580292 | 28.512752 | 20 | 200 |
Visualizations
Here we use some of the data summaries made in the last section to create visual descriptions of the dataset.
Note that ggplot2 is the main graphics
package in R whereas Python primarily uses matplotlib. The two take
different paradigms about creating graphics. Essentially, ggplot2 assumes the
data you’re working with is in long-format whereas matplotlib is easier to work
with if you have already taken the time to do the counting summaries as done
in the last section. However, ggplot2 can handle summarized data with minimal
extra effort, and the seaborn library in Python
makes working with long-format data easier. So, in either language you can work
with whichever format. Examples are presented covering all cases, and one example
is devoted explicitly to showing the differences. We start in the
matplotlib paradigm of plotting summarized data.
Ranking
Recall that there were a couple samples that were seemingly much more mutated than others. A ranking plot will reveal that nicely with the added benefit of showing how the rest of the samples compare as well.
x = np.arange(len(sample_counts))
y = sample_counts.values
plt.plot(y)
plt.scatter(x, y, marker = 'x', s = 50)
plt.title('Mutations per Individual')
plt.xlabel('Individual Rank')
plt.ylabel('Number of Mutations')x = 0:(n_samples-1)
y = unname(sample_counts)
p = ggplot(mapping = aes(x, y)) +
geom_line() +
geom_point(shape = 4) +
ggtitle('Mutations per Individual') +
xlab('Individual Rank') +
ylab('Number of Mutations')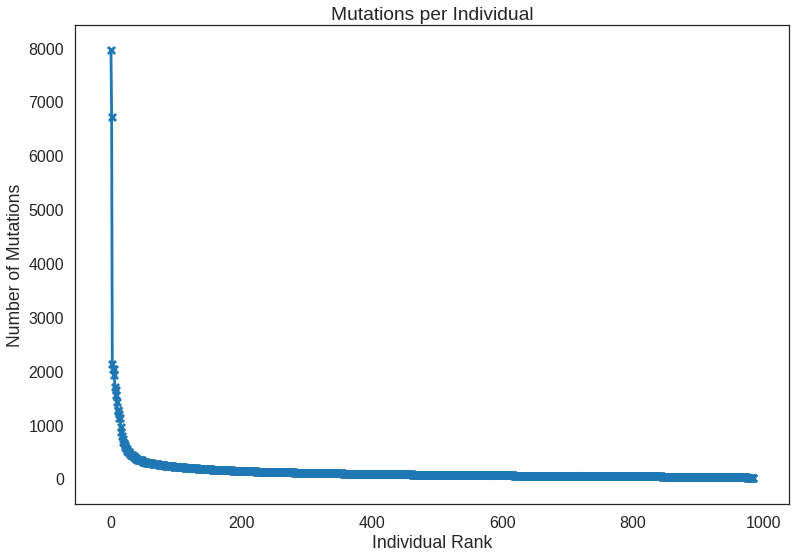
Bar plot
Use the mutation class summary to display all the mutation classes plus how many mutation in each class were found.
plt.barh(
range(len(class_counts)),
class_counts,
tick_label = class_counts.index,
)
plt.gca().invert_yaxis()p = ggplot(
mapping = aes(names(class_counts), unname(class_counts))
) +
geom_bar(stat = 'identity') +
scale_x_discrete(limits = rev(names(class_counts))) +
coord_flip()To achieve the horizontal style, use barh() in Python and coord_flip in R.
Note the plt.gca.invert_yaxis() call needed to get the Python plot to display
in descending order.
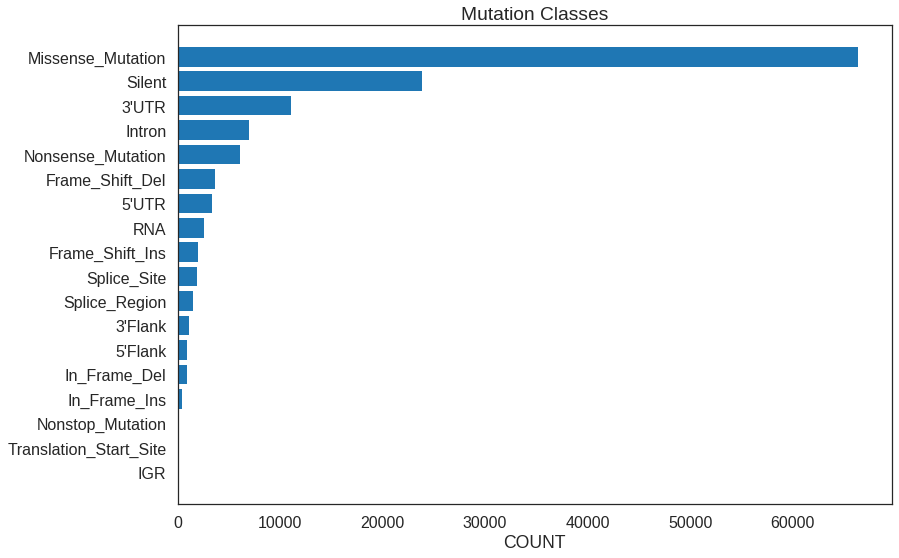
Subplots
Plot two (or more) graphs on the same figure. matplotlib in Python has a built-in
method to handle this, but we’ll need plot_grid() from the cowplot package in R.
fig, (ax1, ax2) = plt.subplots(
1, 2, # 1 row, 2 col
figsize = (20,10)
)
## First plot
# Same as above barplot
ax1.barh(
range(len(class_counts)),
class_counts,
tick_label = class_counts.index,
)
ax1.invert_yaxis()
ax1.set_xlabel('COUNT')
ax1.set_title('Mutation Classes')
## Second plot
n_genes_to_plot = 5
genes = df_summary['GENE'][0:n_genes_to_plot]
counts = df_summary['N_SAMPLES'][0:n_genes_to_plot]
ax2.bar(
np.arange(n_genes_to_plot),
counts,
tick_label = genes
)
ax2.set_xlabel('GENE')
ax2.set_title('Number of Mutated Samples')## First plot
# Reuse instance from above
p1 = p
## Second plot
n_genes_to_plot = 5
genes = df_summary$GENE[0:n_genes_to_plot]
counts = df_summary$N_SAMPLES[0:n_genes_to_plot]
p2 = ggplot(mapping = aes(genes, counts)) +
geom_bar(stat = 'identity') +
scale_x_discrete(limits = genes)
p_grid = plot_grid(p1, p2, ncol = 2)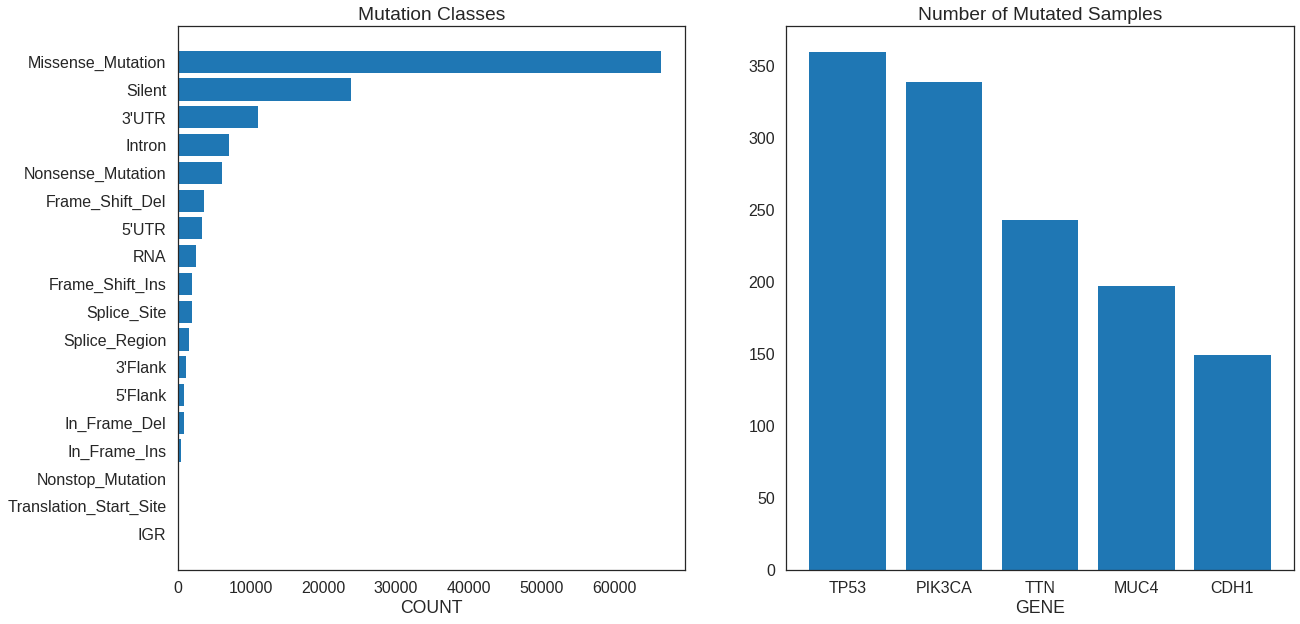
Count vs identity
Thus far we’ve made “identity” plots wherein the numbers plotted were
already calculated. This is the matplotlib paradigm but is easily accomplished
in ggplot2 with the stat = 'identity' option. Without that option, ggplot2
assumes the data is long-format and will do the counting for us. The seaborn
library will let us work with long-format data directly in Python. This example
shows how both methods are used in each language to produce the same output when
counting the number of mutation found in each gene.
## New df's for plotting
df_identity = df_summary.copy()
df_identity.sort_values(
'N_MUTATIONS',
ascending = False,
inplace = True
)
df_count = df.copy()
## Plot params
n_genes_to_plot = 5
genes = df_identity['GENE'][0:n_genes_to_plot]
counts = df_identity['N_MUTATIONS'][0:n_genes_to_plot]
## Get axes
fig, (ax1, ax2) = plt.subplots(
1, 2,
figsize = (20,10)
)
## Identity plot
ax1.bar(
np.arange(n_genes_to_plot),
counts,
tick_label = genes,
color = '#3267AD'
)
ax1.set_title('# Mutations - Identity')
## Count plot
sns.countplot(
x = 'GENE',
data = df[df['GENE'].isin(genes)],
color = '#3267AD',
order = genes,
ax = ax2
)
ax2.set_title('# Mutations - Count')
plt.tight_layout()## New df's for plotting
df_identity = df_summary %>%
arrange(-N_MUTATIONS)
df_count = df
## Plot params
n_genes_to_plot = 5
genes = df_identity$GENE[0:n_genes_to_plot]
counts = df_identity$N_MUTATIONS[0:n_genes_to_plot]
## Identity plot
p1 = ggplot(mapping = aes(genes, counts)) +
geom_bar(stat = 'identity') +
scale_x_discrete(limits = genes) +
ggtitle('# Mutations - Identity')
## Count plot
p2 = ggplot(df, aes(GENE)) +
geom_bar() +
scale_x_discrete(limits = genes) +
ggtitle('# Mutations - Count')
p_grid = plot_grid(p1, p2, ncol = 2)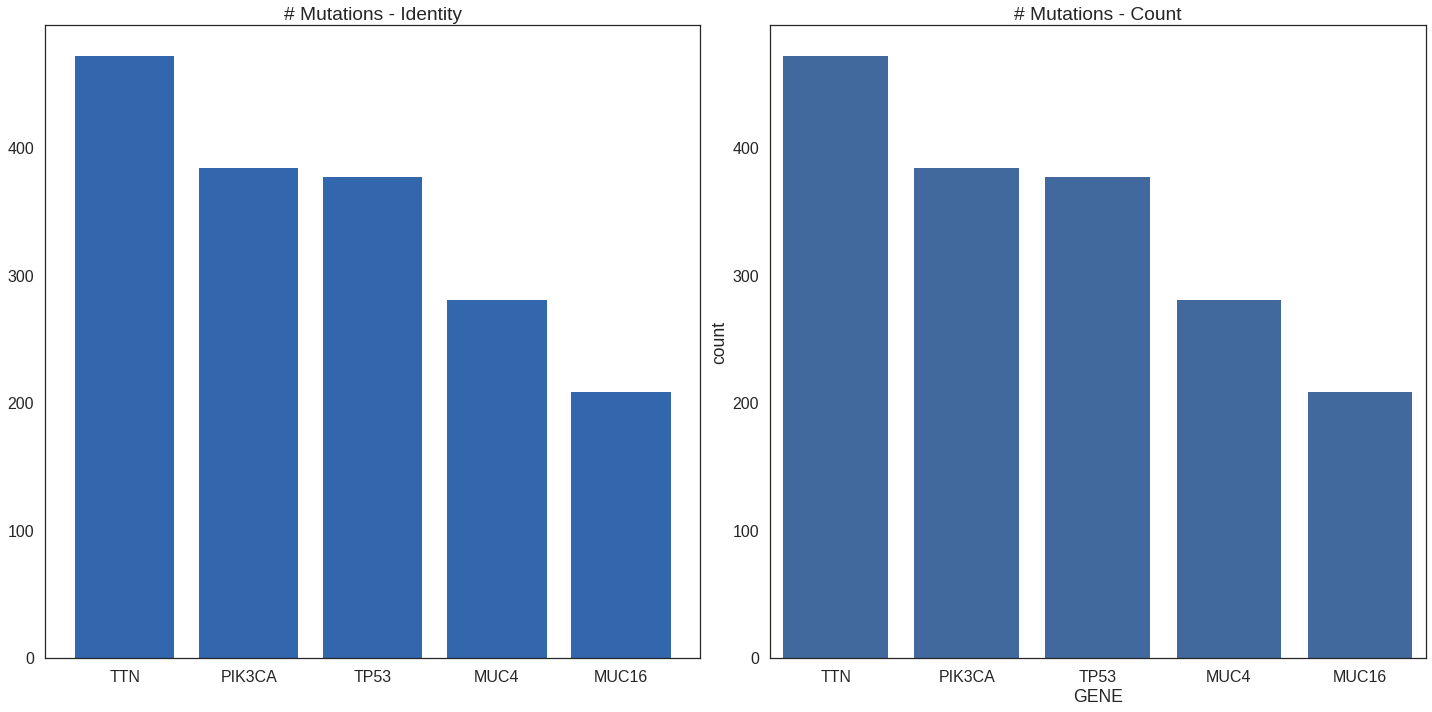
Plot with grouping
In a previous section, we refined our counting procedure to count not just
the number of mutations per gene but rather the number of high-impact, low-impact,
etc. mutations per gene. We can accomplish that graphically as well. We use the
ggplot2 paradigm of passing in the long-format data and letting the plotting packages
do the counting.
Note that here we use a specific color palette (Set1) which exists in both languages.
We could also use the default, another pre-defined palette, or make our own.
## Plot params
n_genes_to_plot = 5
genes = df_summary['GENE'][0:n_genes_to_plot]
## Filter to relevant data
df_plt = df_impact[df_impact['GENE'].isin(genes)]
## Plot
sns.factorplot(
x = 'GENE',
y = 'MUTATION',
hue = 'IMPACT',
order = genes,
palette = 'Set1',
kind = 'bar',
size = 8,
aspect = 2,
data = df_plt
)
plt.title('Impact of Mutations')## Plot params
n_genes_to_plot = 5
genes = df_summary$GENE[0:n_genes_to_plot]
## Filter to relvant data
df_plt = df_impact %>%
filter(GENE %in% genes)
## Reorder for plot
# This is an alternative to
# scale_x_discrete(limits=genes)
df_plt$GENE = factor(df_plt$GENE, levels = genes)
p = ggplot(data = df_plt) +
geom_bar(
aes(x=GENE, y=COUNT, fill=IMPACT),
stat = 'identity',
position = 'dodge'
) +
scale_fill_brewer(
type='qual',
palette = 'Set1'
) +
ggtitle('Impact of Mutations')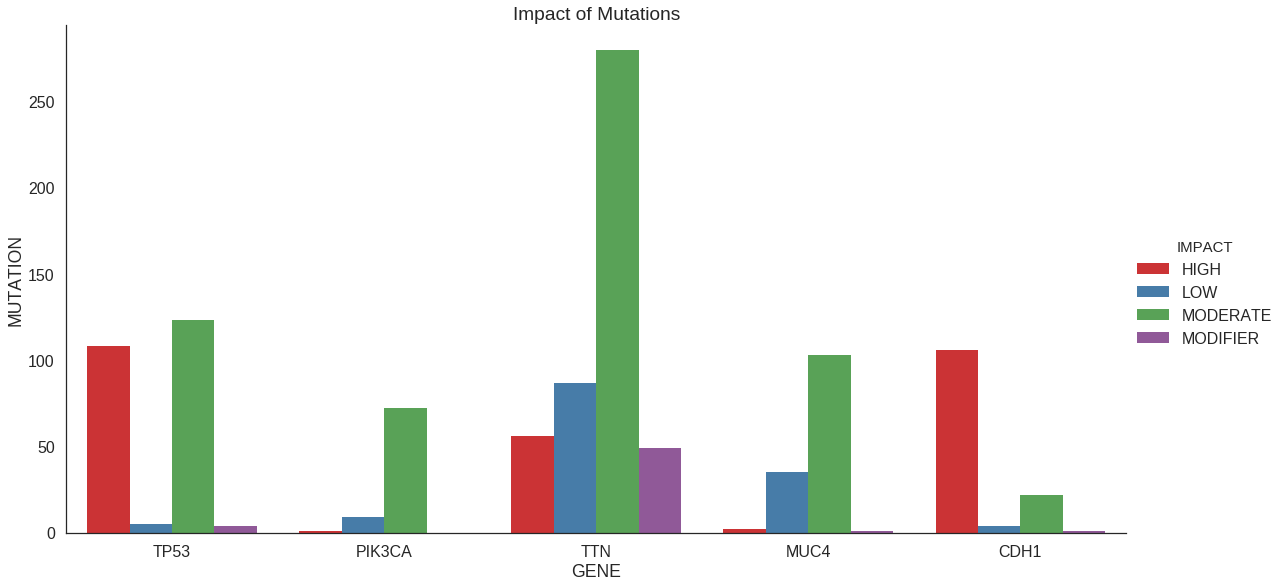
Faceted plot
Let’s do a plot like the one above, but instead look at specific samples rather than aggregated across all samples. This is a good case for a faceted plot.
## Get some samples and genes
samples = sample_counts[0:2].index.tolist()
genes = ['PIK3CA', 'TTN', 'MUC16']
## Filter to relevant data
df_plt = df.loc[(df['SAMPLE'].isin(samples)) & (df['GENE'].isin(genes))]
## Plot
sns.factorplot(
x = 'GENE',
hue = 'IMPACT',
order = genes,
palette = 'Set1',
kind = 'count',
col = 'SAMPLE',
size = 4,
aspect = 1,
data = df_plt
)## Get some samples and genes
samples = names(sample_counts[0:2])
genes = c('PIK3CA', 'TTN', 'MUC16')
## Filter to relevant data
df_plt = df %>%
filter(SAMPLE %in% samples & GENE %in% genes)
## Reorder
df_plt$GENE = factor(df_plt$GENE, levels = genes)
df_plt$SAMPLE = factor(df_plt$SAMPLE, levels = samples)
## Plot
p = ggplot(df_plt) +
geom_bar(
aes(x=GENE, fill=IMPACT),
position = 'dodge'
) +
scale_fill_brewer(
type='qual',
palette = 'Set1'
) +
facet_wrap('SAMPLE')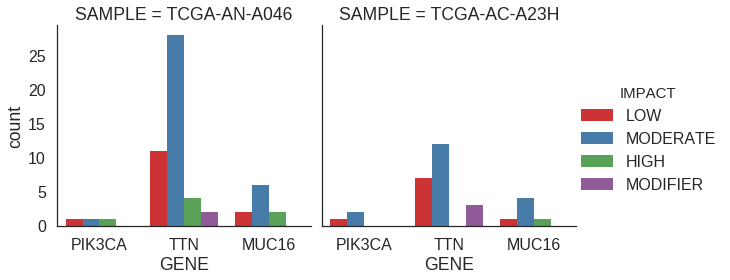
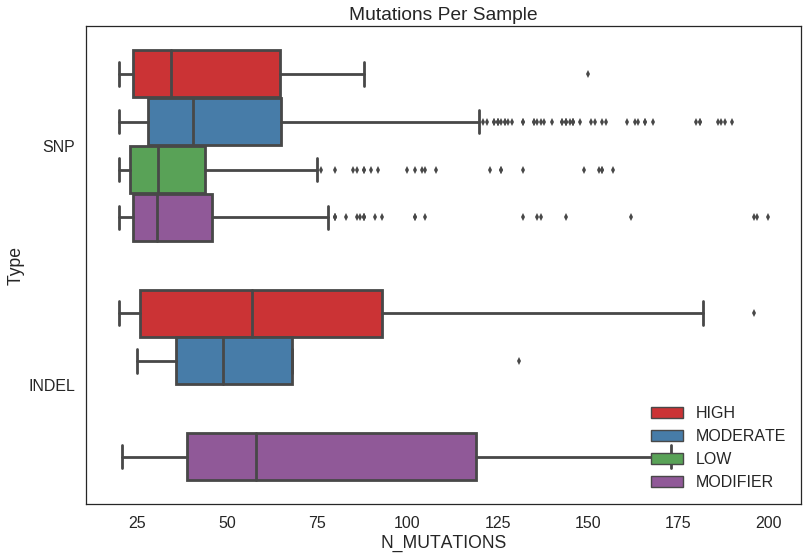
Box plot
sns.boxplot(
y = 'TYPE2',
x = 'N_MUTATIONS',
hue = 'IMPACT',
orient = 'h',
data = df_sample_counts,
order = ['SNP', 'INDEL'],
hue_order = ['HIGH', 'MODERATE', 'LOW', 'MODIFIER'],
palette = 'Set1'
)
plt.title('Mutations Per Sample')
plt.ylabel('Type')
plt.legend(loc=4)## Set order
df_sample_counts$TYPE2 = factor(df_sample_counts$TYPE2, levels = c('INDEL', 'SNP'))
df_sample_counts$IMPACT = factor(df_sample_counts$IMPACT, levels = c(
'HIGH', 'MODERATE', 'LOW', 'MODIFIER'))
## Box plot
p = ggplot(df_sample_counts, aes(TYPE2, N_MUTATIONS, fill = IMPACT)) +
geom_boxplot() +
coord_flip() +
scale_fill_brewer(
type='qual',
palette = 'Set1'
) +
ggtitle('Mutations Per Sample') +bar plot
xlab('Type')